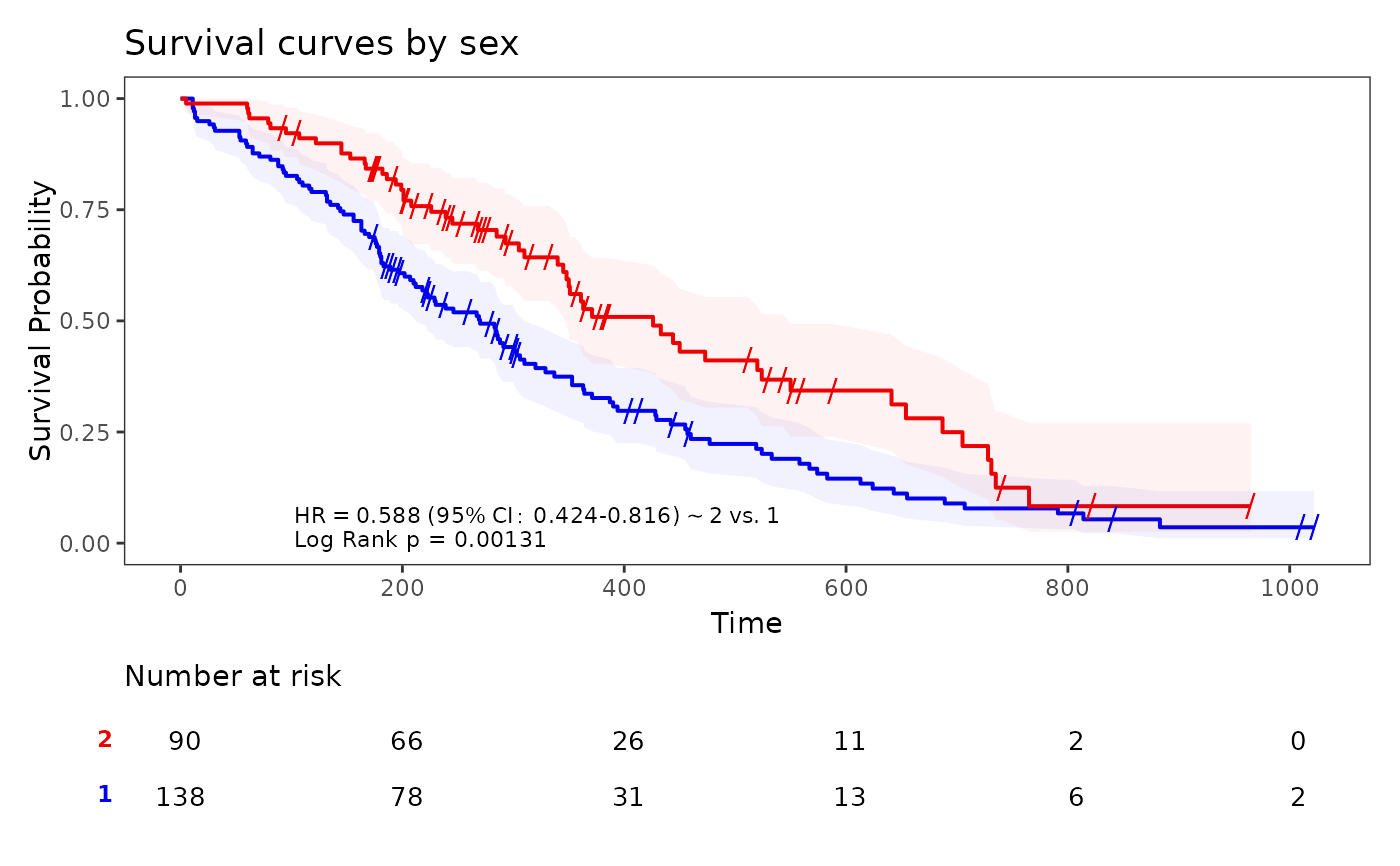
Kaplan-Meier Plots using ggplot
ggkm.RdProduce nicely annotated KM plots using ggplot style.
Usage
ggkm(
sfit,
sfit2 = NULL,
table = TRUE,
returns = TRUE,
marks = TRUE,
CI = TRUE,
line.pattern = NULL,
shading.colors = NULL,
main = "Kaplan-Meier Plot",
xlabs = "Time",
ylabs = "Survival Probability",
xlims = NULL,
ylims = NULL,
ystratalabs = NULL,
cox.ref.grp = NULL,
timeby = 5,
test = c("Log Rank", "Tarone-Ware"),
pval = TRUE,
bold_pval = FALSE,
sig.level = 0.05,
HR = TRUE,
hide_hr_labels = FALSE,
use.firth = 1,
hide.border = FALSE,
expand.scale = TRUE,
legend = FALSE,
legend.xy = NULL,
legend.direction = "horizontal",
line.y.increment = 0.05,
size.plot = 11,
size.summary = 3,
size.table = 3.5,
size.table.labels = 11,
digits = 3,
...
)Arguments
- sfit
an object of class
survfitcontaining one or more survival curves- sfit2
an (optional) second object of class
survfitto compare withsfit- table
logical; if
TRUE(default), the numbers at risk at each time of death is shown as a table underneath the plot- returns
logical; if
TRUEthe plot is returned- marks
logical; if
TRUE(default), censoring marks are shown on survival curves- CI
logical; if
TRUE(default), confidence bands are drawn for survival curves the using cumulative hazard, or log(survival).- line.pattern
linetype for survival curves
- shading.colors
vector of colours for each survival curve
- main
plot title
- xlabs
horizontal axis label
- ylabs
vertical axis label
- xlims
horizontal limits for plot
- ylims
vertical limits for plot
- ystratalabs
labels for the strata being compared in
survfit- cox.ref.grp
indicates reference group for the variable of interest in the cox model. this parameter will be ignored if not applicable, e.g. for continuous variable
- timeby
length of time between consecutive time points spanning the entire range of follow-up. Defaults to 5.
- test
type of test. Either "Log Rank" (default) or "Tarone-Ware".
- pval
logical; if
TRUE(default), the logrank test p-value is shown on the plot- bold_pval
logical; if
TRUE, p-values are bolded if statistically significant atsig.level- sig.level
significance level; default 0.05
- HR
logical; if
TRUE(default), the estimated hazard ratio and its 95% confidence interval will be shown- hide_hr_labels
logical; if
TRUEand there are only two strata, the strata labels are hidden from HR summary because there is only contrast being compared- use.firth
Firth's method for Cox regression is used if the percentage of censored cases exceeds
use.firth. Settinguse.firth = 1(default) means Firth is never used, anduse.firth = -1means Firth is always used.- hide.border
logical; if
TRUE, the upper and right plot borders are not shown.- expand.scale
logical; if
TRUE(default), the scales are expanded by 5% so that the data are placed a slight distance away from the axes- legend
logical; if
TRUE, the legend is overlaid on the graph (instead of on the side).- legend.xy
named vector specifying the x/y position of the legend
- legend.direction
layout of items in legends ("horizontal" (default) or "vertical")
- line.y.increment
how much y should be incremented for each line
- size.plot
text size of main plot
- size.summary
text size of numerical summaries
- size.table
text size of risk table
- size.table.labels
text size of risk table axis labels
- digits
number of digits to round: p-values digits=number of significant digits, HR digits=number of digits after decimal point NOT significant digits
- ...
additional arguments to other methods